For this project you will be studying the p53 protein between many species and building a graph to visualize the relationship between them. This protein is known to be a tumor suppressor and is discussed here: http://www.uniprot.org/uniprot/P04637
0. Obtain the FASTA formatted sequences for the p53 protein from at least 20 species. You can find them here: http://www.ncbi.nlm.nih.gov/protein/?term=p53 and here: http://www.bioinformatics.org/p53/protein.html. An example of the FASTA file for Homo sapiens is show below. We can ignore the FASTA header and just use the sequence that follows it.
>gi|4731632|gb|AAD28535.1|AF135121_1 tumor suppressor protein p53 [Homo sapiens] MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGPDEAPRMPEAA PRVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKMFCQLAKT CPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHERCSDSDGLAPPQHLIRVEGNLRVEYLDDRN TFRHSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGR DRRTEKENLRKKGEPHHELPPGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALEL KDAQAGKEPGGSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD
1. Compute the minimum edit distance between each species’ p53 protein. Use the Needleman-Wunsch implementation that you wrote for hw4. Loop through the >=20 proteins you found. You can put them all in a folder and then loop through all files in that folder. Print out the alignments in a program ProteinCompare.java as follows:
$java ProteinCompare Protein1 Protein1 0 Protein1 Protein2 cost Protein1 Protein3 cost Protein2 Protein2 0 ..
2. Show the minimum alignment between two proteins as shown below. Take the two FASTA files in as input.
$java ShowAlignment p53-AAD28535-homo-sapiens.fa p53-Q95330-rabbit.fa Cost of 61 M M 0 E E 0 E E 0 P S 1 Q Q 0 ... T N 1 E E 0 D D 0 P P 0 G E 1 P 2 D 2 E E 0 A G 1 P L 1 R R 0 M V 1 P P 0 E A 1 A A 0 A P 1 P A 1 R P 1 V E 1 A A 0 P P 0 A A 0 P P 0 A A 0 A A 0 P P 0 T A 1 P L 1 A A 0 A A 0 P P 0 A A 0 P P 0 A A 0 P T 1 S S 0 W W 0 P P 0 ...
3. Write a program Visualize.java to visualize the results using the GraphStream library. Have every species be a node and add an edge between nodes at some threshold of similarity (maybe the mean minimum edit distance). Be sure to have the species name visible.
Deliverables.
Submit your code (ProteinCompare.java, ShowAlignment.java, Visualize.java). In a memo.txt file discuss how varying the gap and mismatch penalty impacts the alignments. Also discuss what relationships you observed in the similarity graph.
Visualize should run as follows using the compile.sh, run.sh, and getclasspath.sh scripts from project 1. You need to include the jars you use in the lib folder and also modify run.sh to call Visualize
$sh compile.sh # compiles files in src into classes folder $sh run.sh # sets the classpath to classes and all the jars in lib then calls Visualize
Grading (total 25 points):
Due: 7/27 @11pm.
5 points: Part 1: ProteinCompare
8 points: Part 2: ShowAlignment
8 points: Part 3: Visualize
4 points: memo.txt and how easy the assignment is to grade
Sample Graphs (made by students)
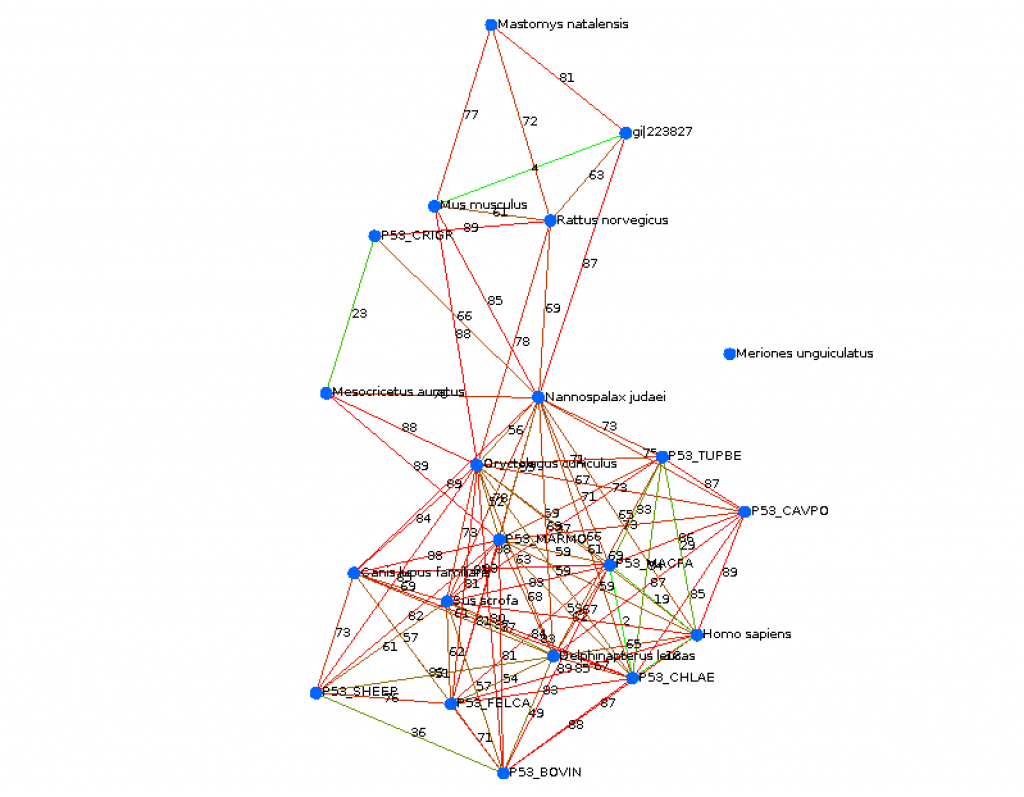
ncadiz (CS310 Summer 2015)
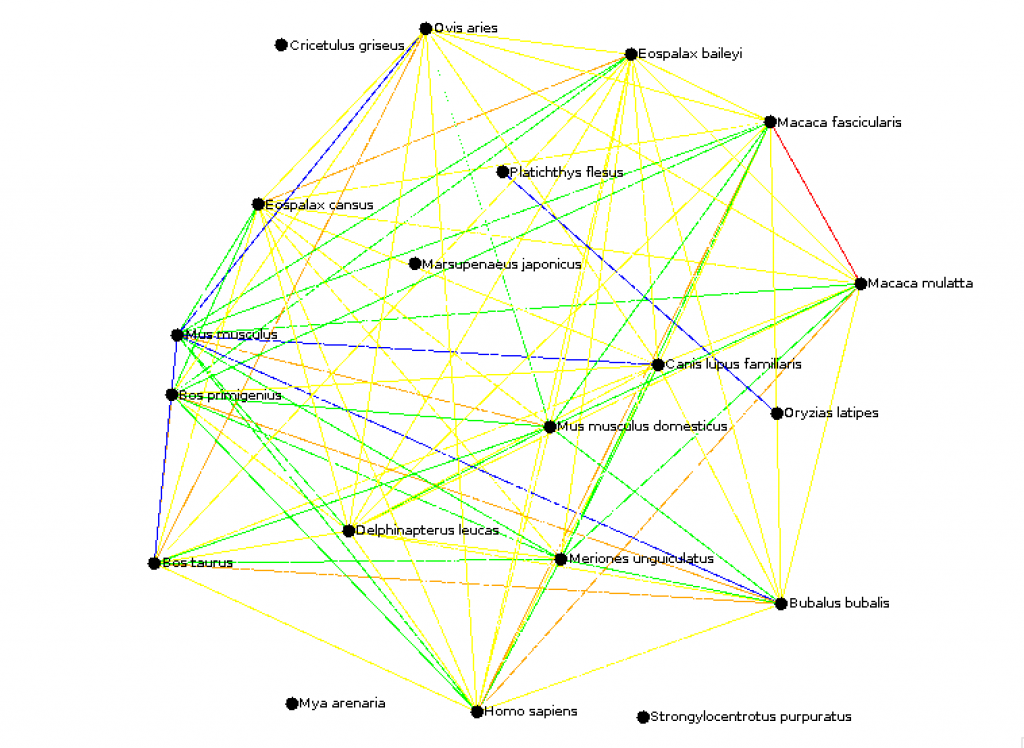
okhan (CS310 Summer 2015) Key: (Mean Percentile goes by the Colors of the Rainbow! Think ROYGBIV) - 00-01% of Mean = Red - 02-20% of Mean = Orange - 21-45% of Mean = Yellow - 46-60% of Mean = Green - 61-100% of Mean = Blue - Outside of Mean = White (Omitted) Mean: 229
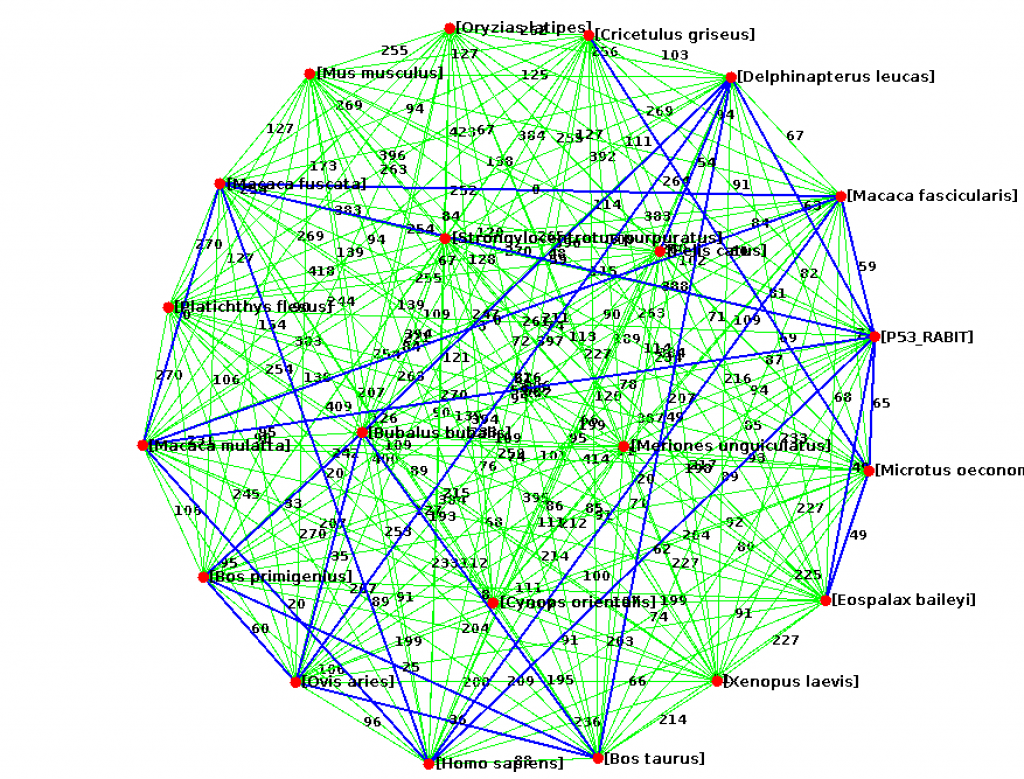
lchen (CS310 Summer 2015) [Xenopus laevis] african-clawed-frog.fasta [Delphinapterus leucas] beluga-whale.fasta [Bos primigenius] bos-primigenius.fasta [Bos taurus] cattle.fasta [Cricetulus griseus] chinese-hamster.fasta [Macaca fascicularis] crab-eating-macaque.fasta [Felis catus] domestic-cat.fasta [Platichthys flesus] european-flounder.fasta [Mus musculus] house-mouse.fasta [Homo sapiens] human.fasta [Macaca fuscata] Japanese-macaque.fasta [Oryzias latipes] japanese-medaka.fasta [Meriones unguiculatus] Mongolian-gerbil.fasta [Cynops orientalis] oriental-fire-bellied-newt.fasta [Eospalax baileyi] plateau-zokor.fasta [P53_RABIT] rabbit.fasta [Strongylocentrotus purpuratus] purple-sea-urchin.fasta [Macaca mulatta] rhesus-monkey.fasta [Microtus oeconomus] Root-vole.fasta [Ovis aries] sheep.fasta [Bubalus bubalis] water-buffalo.fasta
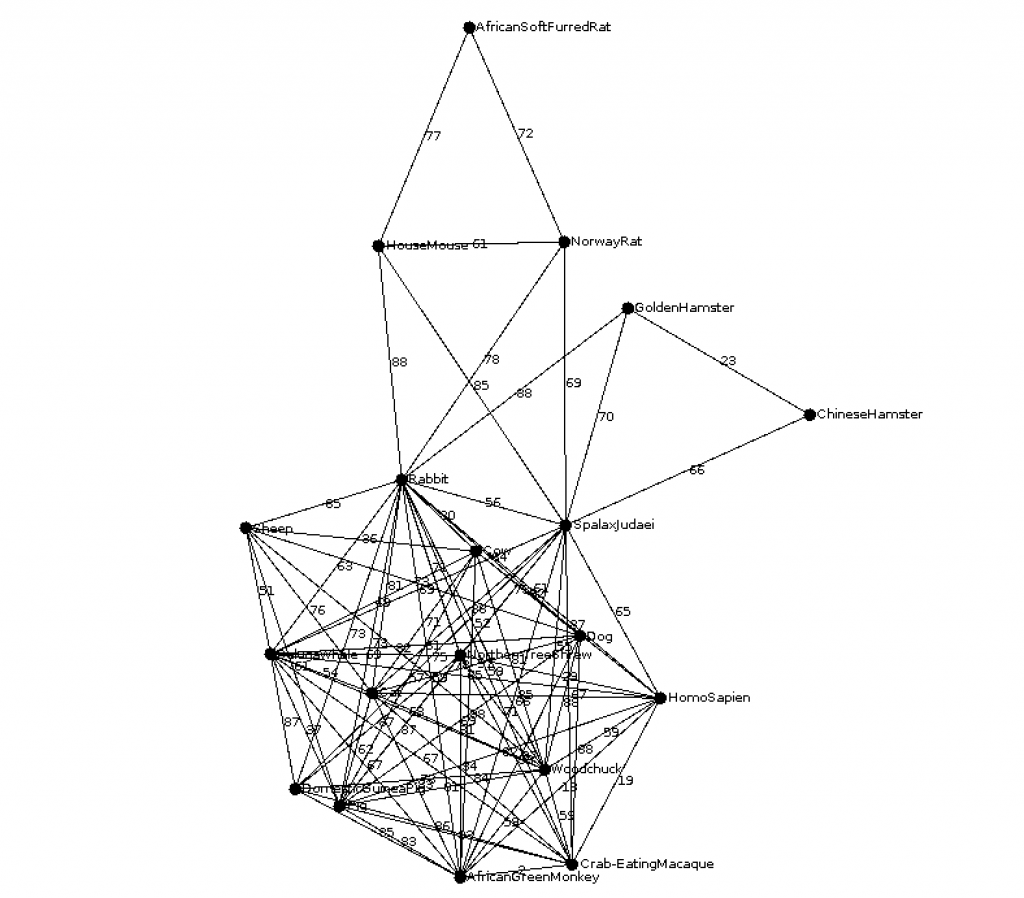
ble002 (CS310 Summer 2015)
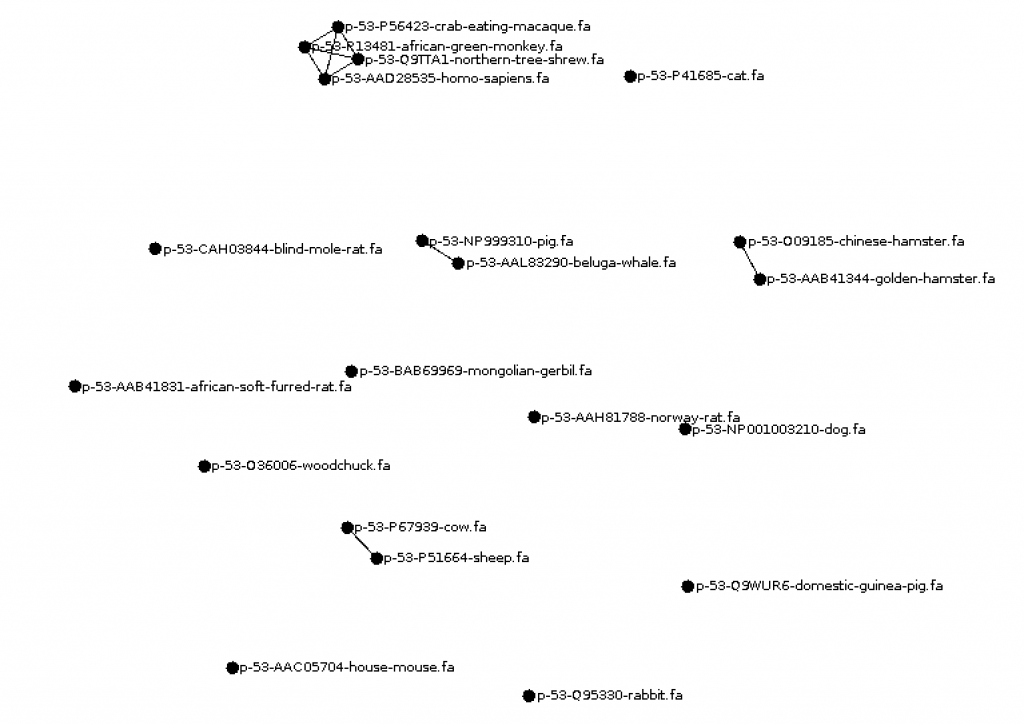
hkim (CS310 Summer 2015)