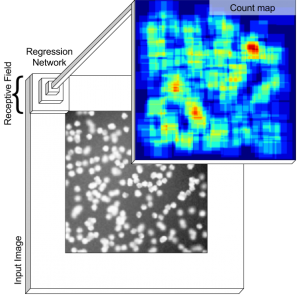
This work develops a method to count heterogeneous objects, such as cell nuclei or sealions. We develop a deep learning based system to that takes as input an image and returns a count of the objects inside and justification for the prediction in the form of weak localization.
Counting objects in digital images is a process that should be replaced by machines. This tedious task is time consuming and prone to errors due to fatigue of human annotators. The goal is to have a system that takes as input an image and returns a count of the objects inside and justification for the prediction in the form of object localization. We repose a problem, originally posed by Lempitsky and Zisserman, to instead predict a count map which contains redundant counts based on the receptive field of a smaller regression network. The regression network predicts a count of the objects that exist inside this frame. By processing the image in a fully convolutional way each pixel is going to be accounted for some number of times, the number of windows which include it, which is the size of each window, (i.e., 32×32 = 1024). To recover the true count we take the average over the redundant predictions. Our contribution is redundant counting instead of predicting a density map in order to average over errors. We also propose a novel deep neural network architecture adapted from the Inception family of networks called the Count-ception network. Together our approach results in a 20% relative improvement (2.9 to 2.3 MAE) over the state of the art method by Xie, Noble, and Zisserman in 2016.
Datasets: The VGG Cells, MBM, and Adipocyte Cells datasets are all freely available here
Code:
| Lasagne + Theano | https://github.com/ieee8023/countception |
| Karas | https://github.com/fizzoo/countception-recreation |
| TensorFlow | https://github.com/rdinse/sea-lion-counter |
| PyTorch | https://github.com/rwightman/pytorch-countception-sealion |
Citation
Cohen, Joseph Paul, et al. “Count-Ception: Counting by Fully Convolutional Redundant Counting.” International Conference on Computer Vision Workshop on BioImage Computing, 2017, http://arxiv.org/abs/1703.08710.