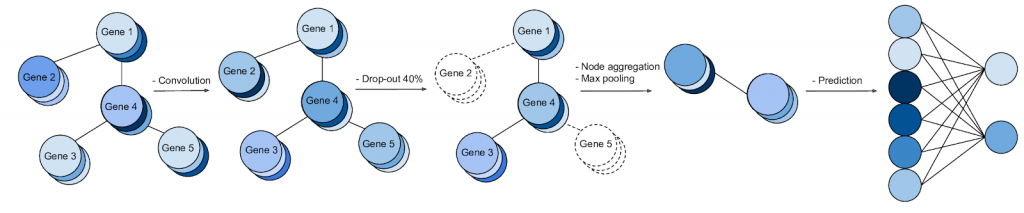
We discuss how gene-gene interaction graphs (same pathway, protein-protein, co-expression, or research paper text association) can be used to impose a bias on a deep neural network model similar to the spatial bias imposed by convolutions on an image. We find this approach provides an advantage for particular tasks in a low data regime but is very dependent on the quality of the graph used. More info: Paper Slides Code
Citation
Francis Dutil, Joseph Paul Cohen, Martin Weiss, Georgy Derevyanko, Yoshua Bengio. “Towards Gene Expression Convolutions Using Gene Interaction Graphs.” International Conference on Machine Learning (ICML) Workshop on Computational Biology (WCB), 2018, http://arxiv.org/abs/1806.06975.
News
Oct, 24th 2018: Martin Weiss received the “Best Presentation Award” for presenting this project at the 2018 IRIC symposium